|
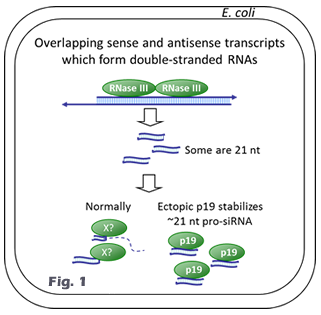
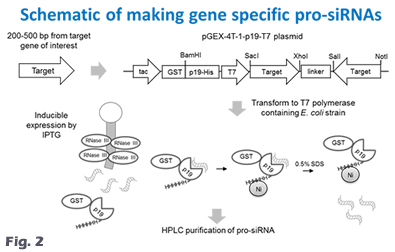
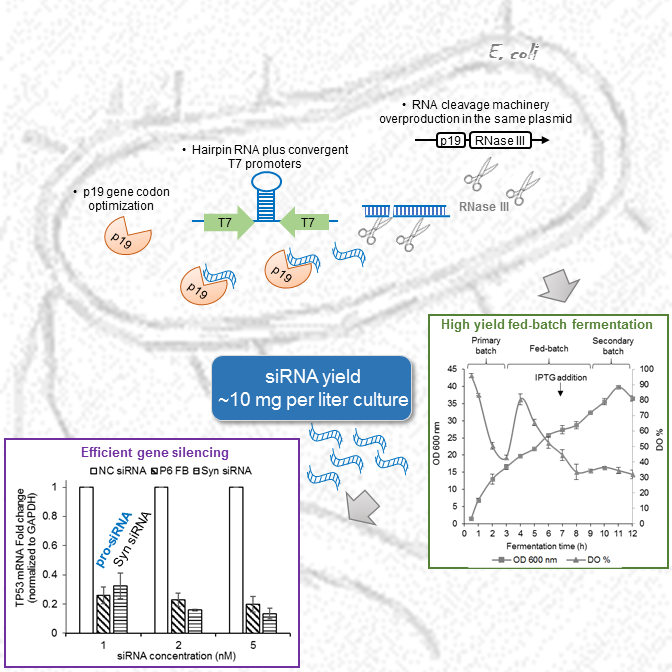
Dr. Linfeng Huang’s research focuses on understanding RNA biology and developing RNA technology and RNA-based precision medicine. He made significant contributions to the RNA interference (RNAi) field including the discovery of the subunit structure of RNA polymerase V (Huang et al., Nature Structural & Molecular Biology, 2009) and the invention of the pro-siRNA (prokaryotic small interfering RNA) technology, which is the world’s first cell-based method for siRNA production (Huang et al., Nature Biotechnology, 2013; Huang & Lieberman, Nature Protocols, 2013). He also co-developed a technique for microRNA detection in live cells (Wang et al., Science Advances, 2020). Currently, Dr. Huang’s lab is developing synthetic biology-based RNA production technology that can be applied to both research and industrial applications. He developed bacterial fermentation methods for large-scale and cost-effective production of siRNAs (Kaur et al., Biotechnology and Bioengineering, 2018). He invented a novel transcriptome-wide RNAi screen method that can be customized to any cells of any species. With an advanced high-throughput screening platform including a high content screening system, his lab has applied high-throughput RNAi screening approach to study important signaling and metabolic pathways (Li et al., BBRC, 2023) and important infectious diseases including bacterial and viral infections. His work has led to the discovery of novel host factors and a chemical inhibitor for Clostridium difficile infection (Li et al., mBio, 2022). He is interested in the basic biology of RNA transcription and regulation, particularly for antisense RNA and double-stranded RNA in bacterial cells (Chan et al., eLife, 2024; Hua et al., mBio, 2022; Huang et al., mBio, 2020). His lab is also investigating the cellular uptake and delivery mechanism of synthetic RNA molecules (Hedlund et al., Nature Communications, 2023; Li et al., BBRC, 2019) and has developed a lipid nanoparticle-based siRNA/cisplatin co-delivery system (Li et al., Chemistry - An Asian Journal, 2019). The overall goal of his research is to identify novel disease-associated genes and to develop potent precision RNA medicines. Our projects utilize a wide range of techniques from biochemical approaches to high-throughput technology. We study a few model organisms from bacteria to mammalian systems. Our lab is also equipped with various state-of-the-art instruments. Research topic 1: pro-siRNA technolgy and RNA biology By serendipity we discovered that ectopic expression of p19 plant viral siRNA binding protein in bacteria stabilizes a ~21 nt siRNA-like small RNAs, named pro-siRNAs (for prokaryotic siRNAs, www.pro-siRNA.com). pro-siRNAs are RNase III degradation products of wide-spread bacterial overlapping sense and antisense transcripts (Fig. 1). And this phenomenon was exploited to make gene specific pro-siRNAs in bacteria which can efficiently and specifically knockdown genes in human cells. The pro-siRNA technology is the world’s first cell-based method of making pure siRNAs (Fig. 2). Research topic 2: RNAi sceening and RNA therapeutics Our lab is continuing to develop pro-siRNA-based technologies for research applications and RNAi therapeutics. We have developed bioprocess-based production method for large scale production of pro-siRNAs. In the future, pro-siRNAs could be used to make personalized RNAi drugs. We have also invented tools for performing transcriptome-wide RNAi screens using the pro-siRNA technology. Our approaches are taking advantage of an in-house high content screening facility. Our tools have been applied to discover important genes involved in human diseases including cancer and infectious diseases. The disease associated genes, that we have discovered, can become therapeutic targets of precision medicines. Our technology has been commercialized by Xiaomo Biotech at Hong Kong Science Park: www.xiaomobio.com Prof. Huang's publications: research page Prof. Huang's bio: bio page
|
|
| Huang lab research data can be downloaded here |